


Solution to Damashek's Riddle

A ROS rich environment is not good for ribosomes


Solution to Damashek's Riddle
WELCOME TO THE DINMAN LAB
For over 30 years, my laboratory studied three distinct yet overlapping fields of study: virology, ribosome structure/function relationships, and regulation of gene expression. I closed my lab in 2022 for a number of reasons. First, because I believe that it is critical to make room for the next generation. Resources such as laboratory space are limited and valuable. Too many researchers hang on for too long. I am proud to have played a central role in establishing the field of translational recoding. Along with my colleagues Phil Farabaugh, Ian Brierley, and Raymond Gesteland/John Atkins, we founded an entirely new field of scientific research. Now it is time to step aside for the next generation. Secondly, during the course of my career, I took on increasingly important administrative roles, e.g. Grad Program Director and Departmental Chair. In November 2025, I took on the role of Director of the Institute for Bioscience and Biotechnology Research (IBBR). I believe that as ones' career progresses, it is important to take on these types of roles so as to serve the greater good of the scientific endeavor.
I am eternally grateful to the people who worked in my lab over the years. You all made wonderful contributions. I am so proud of you.
Regulation of Gene Expression
Since "biological systems tend to use whatever works", there is no reason to believe that programmed ribosomal frameshifting is exclusively utilized by viruses. Based on this philosophy, we are pursuing a bioinformatic program designed to identify functional programmed -1 ribosomal frameshift signals in the genomic databases. This effort employs a combination of computational, DNA microarray, and traditional "wet lab" approaches. We have found that programmed ribosomal frameshift signals can act as mRNA suicide elements, suggesting that PRF is used to post-transcriptionally regulate the abundance of specific mRNAs and their encoded protein products. The reverse side of this coin is the question of how viruses have evolved to circumvent this regulatory mechanism, allowing them to utilize programmed ribosomal frameshifting without having their mRNAs degraded.


Virology
The maintenance of correct translational reading frame is fundamental to the integrity of the protein synthetic process, and ultimately to cell growth and viability. Despite this, it has been demonstrated that certain viruses utilize specific signals on their mRNAs that induce elongating ribosomes to shift reading frame. The highly controlled efficiencies of PRF events ensure that the proper stoichiometric ratio of viral structural to enzymatic proteins are available for viral particle assembly. Changing frameshifting efficiencies alters this ratio, preventing proper viral particle assembly and interfering with virus propagation. Thus, programmed ribosomal frameshifting presents a promising new target for anti-viral pharmacological intervention. We are characterizing a series of yeast mutants and drugs in order to identify new targets for antiviral therapies. We are also working to create a reverse genetic system for a dsRNA virus of yeast.
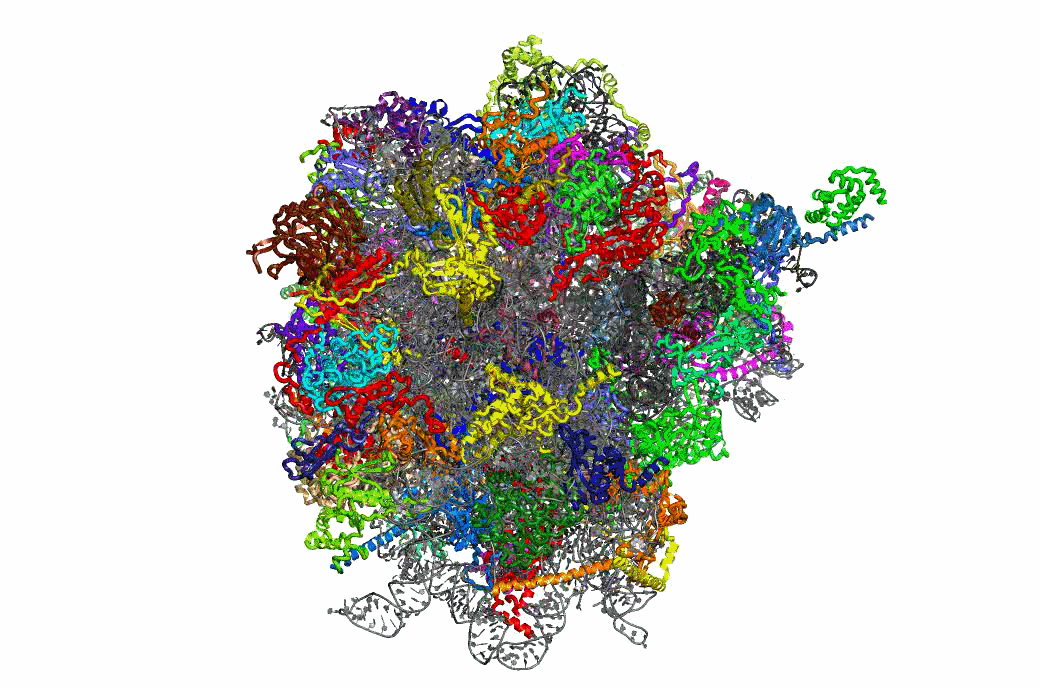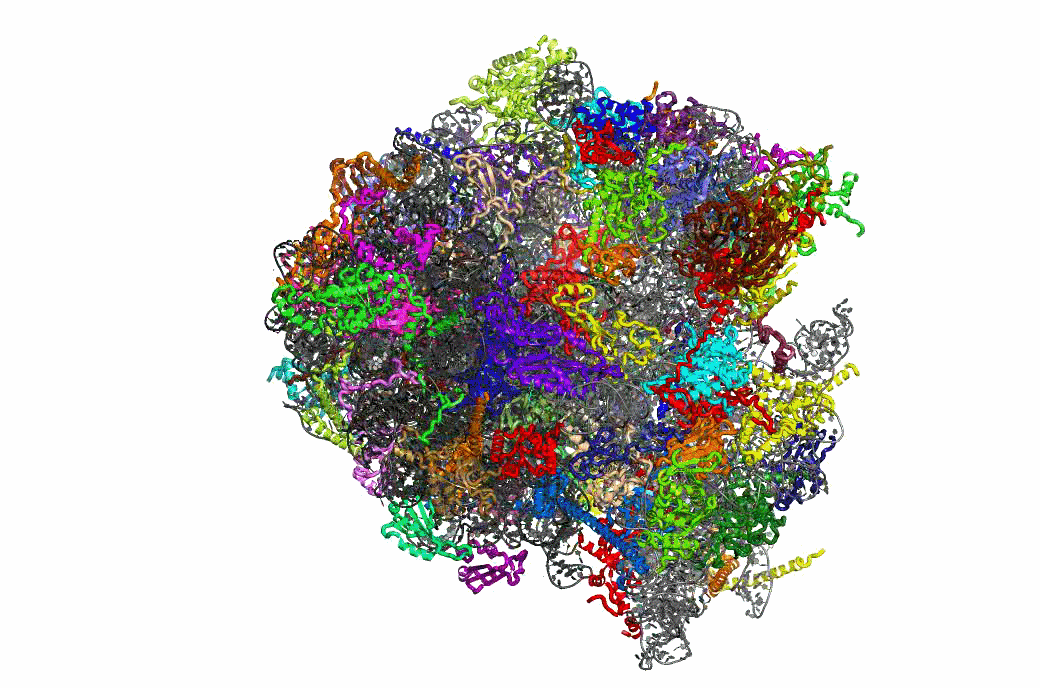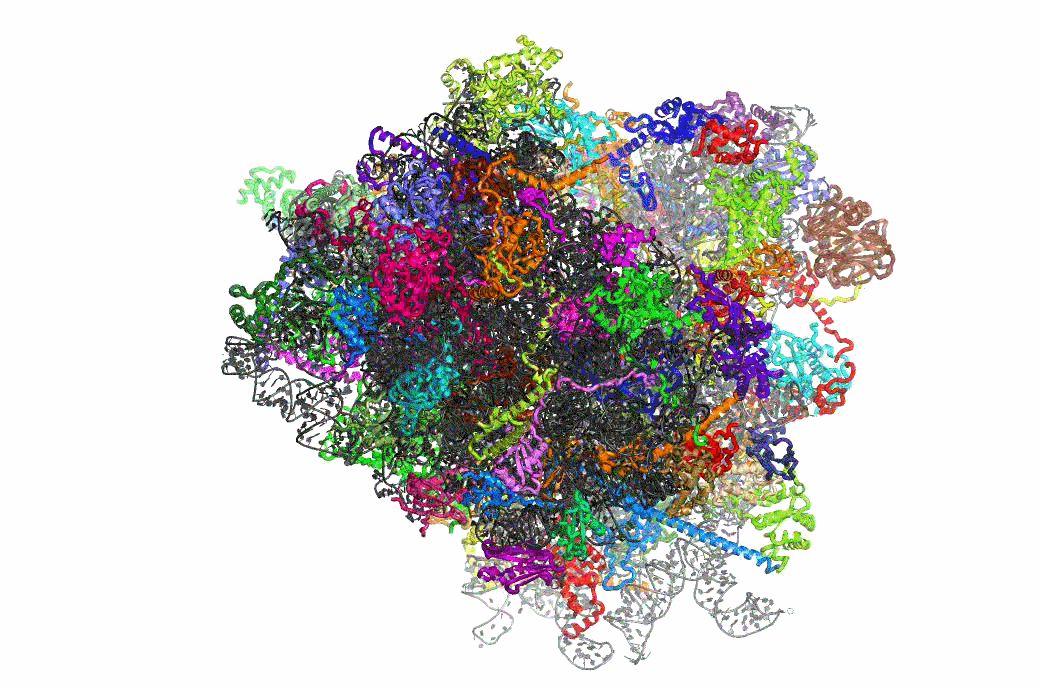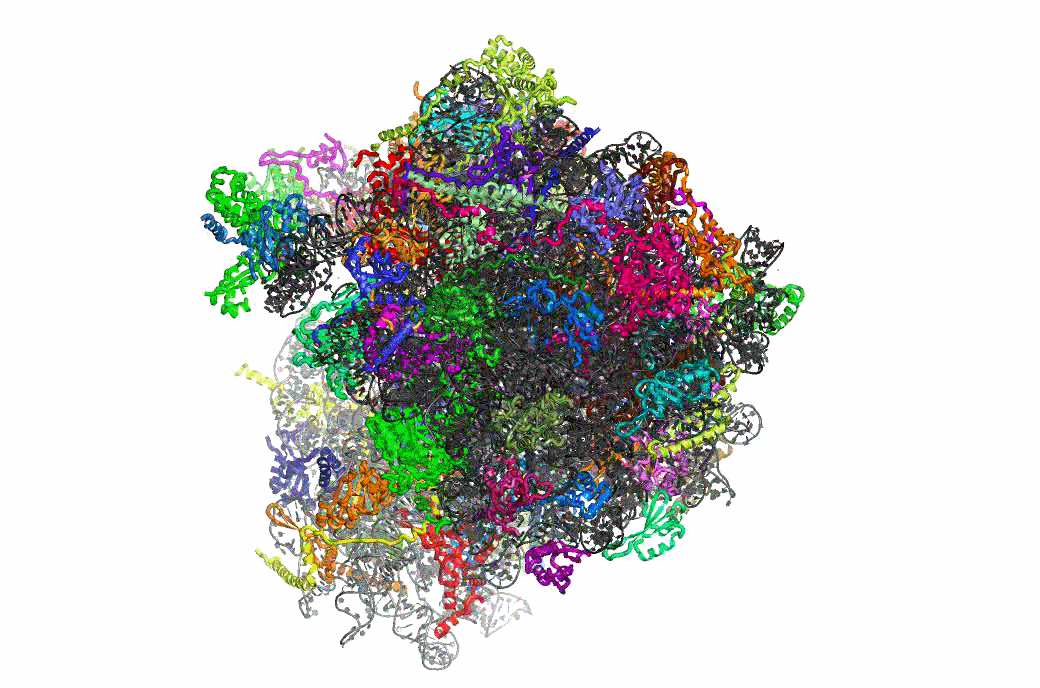
Ribosome Structure & Function
One important function of the ribosome is to faithfully maintain translational reading frame. Viral mRNA signals that abrogate this function by programming ribosomes to shift frame have proved to be of tremendous utility in elucidating the molecular mechanisms underlying this essential task. The newly available atomic resolution structures of ribosomes mark a critical milestone in the quest to link ribosome structure with function, and our studies on PRF have begun to link ribosome structure with translational frame maintenance. We have shown that both the biophysical interactions between ribosomal proteins rRNAs and tRNAs, and the biochemical properties of ribosome-associated enzymatic activities are both important for proper reading frame maintenance. On a broader scale, our work also is consistent with the hypothesis that communication between the different functional centers of the ribosome is critical for coordinating ribosome structure with its various functions. Of particular interest, recent structural analyses of mutants that we had previously identified as affecting frameshifting reveals that they correspond to critical points of contact between specific ribosomal components. This positions us for to conduct reverse genetic studies linking ribosome structure with function.
CONTACT US
4062 Campus Drive
College Park MD 20742
Office: 301 405 0918
Lab: 301 405 1758
FAX: 301 314 9489
email: dinman@umd.edu
Useful Links:
| UMD Home | Directories | Search | Admissions | UMD Calendar |
| College of Computer Mathematical and Natural Sciences | CBMG
